GFF to BED conversion
It exists many GFF formats and many GTF formats (see here for a complete review) and many tools to perform the conversion. We will try to see in this review the main differences.
Table of Contents
Test summary
| tool | Comment |
|---|---|
| AGAT | default RGB color to 255,0,0 |
| PASA | Particular 3rd column that contains a list of names |
| bedops | each gff feature give one line. Only the 6 first colums are correct |
| Kent utils | extra coma at the end of 11th and 12th column |
The GFF file to convert
The test file is a GFF3 file:
##gff-version 3
# This is a test sample
scaffold625 maker gene 337818 343277 . + . ID=CLUHARG00000005458;Name=TUBB3_2
scaffold625 maker mRNA 337818 343277 . + . ID=CLUHART00000008717;Parent=CLUHARG00000005458
scaffold625 maker tss 337916 337918 . + . ID=CLUHART00000008717:tss;Parent=CLUHART00000008717
scaffold625 maker start_codon 337916 337918 . + . ID=CLUHART00000008717:start;Parent=CLUHART00000008717
scaffold625 maker CDS 337915 337971 . + 0 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 maker CDS 340733 340841 . + 0 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 maker CDS 341518 341628 . + 2 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 maker CDS 341964 343033 . + 2 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 maker stop_codon 343031 343033 . + . ID=CLUHART00000008717:stop;Parent=CLUHART00000008717
scaffold625 maker exon 337818 337971 . + . ID=CLUHART00000008717:exon1;Parent=CLUHART00000008717
scaffold625 maker exon 340733 340841 . + . ID=CLUHART00000008717:exon2;Parent=CLUHART00000008717
scaffold625 maker exon 341518 341628 . + . ID=CLUHART00000008717:exon3;Parent=CLUHART00000008717
scaffold625 maker exon 341964 343277 . + . ID=CLUHART00000008717:exon4;Parent=CLUHART00000008717
scaffold625 maker five_prime_utr 337818 337914 . + . ID=CLUHART00000008717:five_prime_utr;Parent=CLUHART00000008717
scaffold625 maker three_prime_UTR 343034 343277 . + . ID=CLUHART00000008717:three_prime_utr;Parent=CLUHART00000008717
AGAT
AGAT v0.2.2
agat_convert_sp_gff2bed.pl --gff 1_test.gff -o 1_test_agat.bed
scaffold625 337817 343277 CLUHART00000008717 0 + 337914 343033 255,0,0 4 154,109,111,1314 0,2915,3700,4146
PASA
PASA pasa-v2.4.1
./PASApipeline/misc_utilities/gff3_file_to_bed.pl test_1.gff > 1_test_transdecoder.bed
#gffTags
scaffold625 337817 343277 ID=CLUHART00000008717;CLUHARG00000005458;TUBB3_2 0 + 337914 343033 0 4 154,109,111,1314 0,2915,3700,4146
bedops
version: 2.4.37
gff2bed < 1_test.gff > 1_test_bedops.bed
scaffold625 337817 337914 CLUHART00000008717:five_prime_utr . + maker five_prime_utr . ID=CLUHART00000008717:five_prime_utr;Parent=CLUHART00000008717
scaffold625 337817 337971 CLUHART00000008717:exon1 . + maker exon . ID=CLUHART00000008717:exon1;Parent=CLUHART00000008717
scaffold625 337817 343277 CLUHARG00000005458 . + maker gene . ID=CLUHARG00000005458;Name=TUBB3_2
scaffold625 337817 343277 CLUHART00000008717 . + maker mRNA . ID=CLUHART00000008717;Parent=CLUHARG00000005458
scaffold625 337914 337971 CLUHART00000008717:cds . + maker CDS 0 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 337915 337918 CLUHART00000008717:start . + maker start_codon . ID=CLUHART00000008717:start;Parent=CLUHART00000008717
scaffold625 337915 337918 CLUHART00000008717:tss . + maker tss . ID=CLUHART00000008717:tss;Parent=CLUHART00000008717
scaffold625 340732 340841 CLUHART00000008717:cds . + maker CDS 0 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 340732 340841 CLUHART00000008717:exon2 . + maker exon . ID=CLUHART00000008717:exon2;Parent=CLUHART00000008717
scaffold625 341517 341628 CLUHART00000008717:cds . + maker CDS 2 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 341517 341628 CLUHART00000008717:exon3 . + maker exon . ID=CLUHART00000008717:exon3;Parent=CLUHART00000008717
scaffold625 341963 343033 CLUHART00000008717:cds . + maker CDS 2 ID=CLUHART00000008717:cds;Parent=CLUHART00000008717
scaffold625 341963 343277 CLUHART00000008717:exon4 . + maker exon . ID=CLUHART00000008717:exon4;Parent=CLUHART00000008717
scaffold625 343030 343033 CLUHART00000008717:stop . + maker stop_codon . ID=CLUHART00000008717:stop;Parent=CLUHART00000008717
scaffold625 343033 343277 CLUHART00000008717:three_prime_utr . + maker three_prime_UTR . ID=CLUHART00000008717:three_prime_utr;Parent=CLUHART00000008717
Kent utils
version from 26-Feb-2020
./gff3ToGenePred.dms 1_test.gff temp.genePred
./genePredToBed.dms temp.genePred 1_test_genePred.bed
scaffold625 337817 343277 CLUHART00000008717 0 + 337914 343033 0 4 154,109,111,1314, 0,2915,3700,4146,
The bed format
Detailed information can be found here: https://genome.ucsc.edu/FAQ/FAQformat.html
Below a description of the different fields:
| column | feature type | mandatory | comment |
|---|---|---|---|
| 1 | chrom | X | The name of the chromosome (e.g. chr3, chrY, chr2_random) or scaffold (e.g. scaffold10671). |
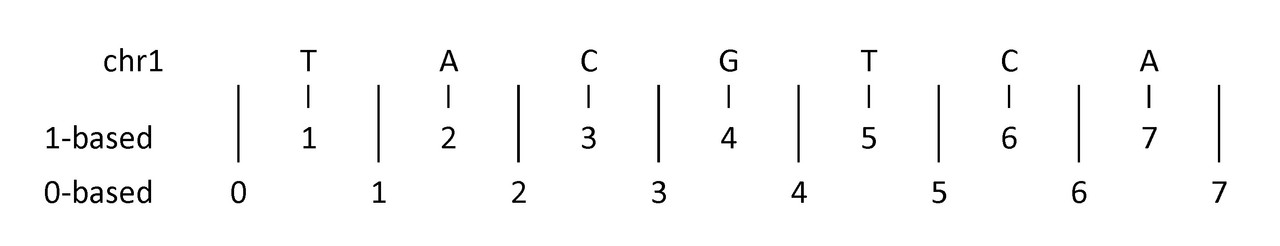
| 2 | chromStart | X | The starting position of the feature in the chromosome or scaffold. The first base in a chromosome is numbered 0. |
| 3 | chromEnd | X | The ending position of the feature in the chromosome or scaffold. The chromEnd base is not included in the display of the feature. For example, the first 100 bases of a chromosome are defined as chromStart=0, chromEnd=100, and span the bases numbered 0-99. |
| 4 | name | Defines the name of the BED line. This label is displayed to the left of the BED line in the Genome Browser window when the track is open to full display mode or directly to the left of the item in pack mode. | |
| 5 | score | A score between 0 and 1000. If the track line useScore attribute is set to 1 for this annotation data set, the score value will determine the level of gray in which this feature is displayed (higher numbers = darker gray). | |
| 6 | strand | Defines the strand - either '+' or '-'. | |
| 7 | thickStart | The starting position at which the feature is drawn thickly | |
| 8 | thickEnd | The ending position at which the feature is drawn thickly | |
| 9 | itemRgb | An RGB value of the form R,G,B (e.g. 255,0,0). If the track line itemRgb attribute is set to "On", this RBG value will determine the display color of the data contained in this BED line. NOTE: It is recommended that a simple color scheme (eight colors or less) be used with this attribute to avoid overwhelming the color resources of the Genome Browser and your Internet browser. | |
| 10 | blockCount | The number of blocks (exons) in the BED line. | |
| 11 | blockSizes | A comma-separated list of the block sizes. The number of items in this list should correspond to blockCount. | |
| 12 | blockStarts | A comma-separated list of block starts. All of the blockStart positions should be calculated relative to chromStart. The number of items in this list should correspond to blockCount. |
/!\ location BED format is 0-based, half-open [start-1, end), while GFF is 1-based, closed [start, end].